![]()
![]()
![]()
Use LEFT and RIGHT arrow keys to navigate between flashcards;
Use UP and DOWN arrow keys to flip the card;
H to show hint;
A reads text to speech;
235 Cards in this Set
- Front
- Back
|
DRAW CELL CYCLE AND KNOW CELLULAR FUNCTIONS AT EACH PHASE |
|
|
|
RELATIONSHIP BTWN LIFESPAN OF CELL & DISEASE |
Long lived cells can accumulate products or toxic damage over time that can result in cell death e.g. Alzheimer's
|
|
|
RELATIONSHIP BTWN RATE OF CELL TURNOVER & DISEASE |
Cells w/rapid turnover have increased chance of accumulating mutations that affect growth & division, leading to diseases e.g. cancer |
|
|
HIERARCHY OF STEM CELLS |
Totipotent (can give rise to any of the cell types) ---> Pluripotent (can give rise to multiple cell types) --> Blood Stem Cells (can give rise to RBC or WBC) or Other Stem Cells (can give rise to Muscle, Nerve, Bone & Other tissues)
*As you go further down the hierarchy, the stem cells are able to give rise to less differentiated cell types
*Induced Pluripotent Stem Cells - We can reprogram stem cells and essentially get them to go backwards |
|
|
CRITERIA FOR CELL DEATH |
1. Plasma membrane breakdown 2. Cell explodes 3. Cell eaten by other cells |
|
|
CELL DEATH: PROPOSED POINTS OF NO RETURN |
1) Massive activation of caspases 2) Protracted loss of membrane potential 3) Phosphatidyl serine (PS) exposure which triggers for phagocytosis -- normally PS is on the inner leaflet |
|
|
4 TYPES OF CELL DEATH |
1. APOPTOSIS - Cells killing themselves 2. AUTOPHAGY - Cell digesting own organelles; Results in vacuolization of the cytoplasm 3. CORNIFICATION - Commonly occurs in skin; involves removal of cytoplasmic content - cell is dead but retains its shape 4. NECROSIS - Basically all other forms of cell death |
|
|
APOPTOSIS: MORPHOLOGICAL FEATURES |
*Cytoplasmic shrinking *Rounding up of the cell *Retraction of pseudopodes *Nuclear shrinkage (pyknosis) *Membrane blebbing *Nuclear fragmentation (karyorrhexis) -- [aka Intranucleosomal DNA Fragmentation] - DNA cuts up into little pieces; ordered disassembly from inside *Engulfment by phagocytes/macrophages -Minor modification of cytoplasmic organelles *Phosphatidyl serine (PS) flipping to extracellular portion of the Plasma Membrane - exposure of PS to outside of cell MARKS cell for rapid consumption by Macrophages *Cell loses attachment to substrata it was growing on *Cell shrivels up (without lysing)
|
|
|
AUTOPHAGY: MORPHOLOGICAL FEATURES |
-Lack of chromatin condensation -Massive vacuolization of the cytoplasm -Accumulation of double membraned autophagic vacuoles -Little or no uptake by phagocytic cells |
|
|
CORNIFICATION: MORPHOLOGICAL FEATURES |
-Elimination of cytosolic organelles -Accumulation of lipids -Extrusion of lipids in extracellular space -Modifications of plasma membrane -Desquamation (loss of corneocytes) due to protease activation |
|
|
NECROSIS: MORPHOLOGICAL FEATURES |
-Cytoplasmic swelling (oncosis) -Rupture of plasma membrane -Swelling of cytoplasmic organelles -Moderate chromatin condensation |
|
|
ERYTHROPOIETIN |
*Key protein involved in regulating rate of Red Blood Cell Production (aka erythropoiesis)
*Thus, it is involved in regulating # of RBC |
|
|
RED BLOOD CELL PRODUCTION (aka ERYTHROPOIESIS) |
*Triggered by Hypoxia (lack of adequate oxygen supply) occurs due to decreased RBC count, decreased availabity of Oxygen to blood, or increased tissue demands for Oxygen *Results in reduced Oxygen levels in blood *Kidney releases erythropoietin *Erythropoeitin stimulates Red Bone Marrow *Enhanced erythropoiesis occurs *Results in more RBC *Increased Oxygen carrying ability of blood *Return to Normal Blood Oxygen levels |
|
|
LIFESPAN OF RBC |
120 DAYS |
|
|
ERYTHROPOIESIS - BIRTH & CELLULAR STAGES |
*Process takes 7 days *Begins with proerythroblast --> Basophilic erythroblast --> Polychromatophilic erythroblast --> Orthochromatophilic erythroblast --> Reticulocyte --> Erythrocyte
*Reticulocyte's can be used as diagnostic tool: High amt of Reticulocytes = Hemolytic Anemia b/c means more red blood cells are being made by the bone marrow. This can occur after a lot of bleeding, a move to a high altitude, or Hemolytic Anemia, condition in which RBC are being broken down (hemolysis) |
|
|
ERYPTOSIS |
Programmed cell death for the erythrocyte - As erythrocyte ages, changes in membrane composition (i.e. PS on OUTER LEAFLET) cause it to be recognized by macrophages in spleen/liver/bone marrow *Flipase is the enzyme that flips PS to the inner leaflet - this process requires NRG...so when there's not enough NRG provided, PS accumulates on outer leaflet and then phagocytosed - Old cells therefore removed by phagocytosis |
|
|
SICKLE CELL ANEMIA |
- Caused by mutations in the beta-globin gene; occurs in ~1:500 African-American children
SIGNS OF SICKLE CELL ANEMIA: - Sickle cell crisis (5-7 days) - Anemia - Vaso-occlusion (ischemic even to organs) - Aplastic anemia (viral attack on RBC production) - Hemolytic crisis (increased RBC breakdown, G6PD) - Sequestration (splenic infarction) |
|
|
NEURONS IN THE CORTEX SERVE AS AN EXAMPLE OF WHAT? |
- Long lived cells
- An instance where cell differentiation is linked to cell cycle *There are 3 types of Neuronal Precursors *Way in which cell develops fr the precursor all depends on cell cycle: a. Proliferative division b. Differentiative asymmetrical division - results in 1 neuron cell from a precursor c. Differentiative symmetrical division - results in 2 neuron cells from precursors
|
|
|
NEURONAL DEATH |
1. Acute changes in neuronal environment that induce cell death (ischemia & apoptosis) 2. Trauma (i.e. gunshot) 3. Chronic changes that affect neuron indirectly (immune mediated demyelination, e.g. multiple sclerosis) 4. Protein aggregates & neuronal degeneration (when a protein misfold & forms aggregate) |
|
|
ALZHEIMER'S DISEASE |
*most common neurological disorder *predicted to affect 1/80 globally by 2050 *mostly affects ppl over age 65 *considered both a protein aggregate disease and a tauopathy *Amyloid Plaques & Neurofibrilliary Tangles (NFT) -- Hallmarks of Alzheimer's *Associated with beta-amyloid aggregates |
|
|
LIFESPAN OF A PROTEIN |
*Proteins, like cells, also degrade *Half life of proteins ranges fr 0.2 hrs to years *Proteins can be non-specifically turned over (degraded) by mechanisms such as PROTEOLYSIS *Proteins can be specifically targeted for degradation by UBIQUITINATION *There is a regulated balance btwn synthesis of proteins & destruction |
|
|
DEATH BY UBIQUITINATION |
*Ubiquitin - serves as the tag that marks a protein for degradation
*The Ubiquitin-Proteasome Pathway* Ub itself does not degrade proteins. It serves only as a tag that marks proteins for degradation. -The degradation itself is carried out by a proteasome - Proteins that are to be degraded are first tagged by conjugating them with Ub and these tagged proteins are then recognized and shuttled to the proteasome for degradation - Involves interaction w/3 enzymes
THINK OF PROTEASOME AS THE CELL'S GARBAGE CAN
Cell cycle & Circadian Rhythm depend on ubiquitination |
|
|
LONG LIVED PROTEINS |
- Certain proteins turn over v. slowly or never turn over/get modified once formed - e.g. elastin in the skin endure UV damage, resulting in wrinkles - another e.g. crystallines in the eyes' lens, results in cataracts - Hence diseases relating to those proteins are AGING DISORDERS (and occur due to accumulated damage)
|
|
|
DEVELOPMENT OF THE HUMAN CORTEX |
|
|
|
KEY TAKEAWAY POINTS FROM LAMBERT'S CELL CYCLE SLIDES |
1. Cells arise from a proliferating precursor regulated by cell cycle 2. Cell death can be programmed, or happen as a result of aging/accumulation of damage 3. Lifespan of cells within the body can vary greatly 4. Balance btwn birth & death of cells MUST be maintained 5. Longer living cells manifest problems associated w/accumulation of damage (aging) 6. Creation & Destruction of cellular Proteins has similar patterns (to creation & destruction of cells) |
|
|
ISCHEMIA/REPERFUSION |
Injury arising from acute myocardial infarction, cardiac surgery, and chronic heart failure
*Infarction - Tissue death caused by local lack of Oxygen due to obstruction of tissue's blood supply **Myocardial Infarction is the partial death of heart tissue (aka heart attack) |
|
|
TOXIC XENOBIOTICS (EXAMPLES) |
-Arsenic -Heavy Metals (Lead, Mercury, etc) -Salicylate (FYI: salicylate is a ubiquitous agent, used as an analgesic agent for the treatment of mild to moderate pain, common sources of unintentional and suicidal ingestion) -Acetaminophen |
|
|
NEURODEGENERATIVE DISEASES (EXAMPLES) |
*Alzheimer's - (AD) most common form of dementia; Know the Amyloid Hypothesis *Parkinson's - (PD) degenerative disorder of the central nervous system *Huntington's - (HD) a genetic neuro-degenerative disorder affecting muscle coordination and leads to cognitive decline and behavioral symptoms |
|
|
Alzheimer's Disease: The Amyloid Hypothesis |
The amyloid hypothesis postulates that extracellular beta-amyloid (Aβ) deposits are the fundamental cause of the disease |
|
|
APOPTOSIS: FACTS AT A GLANCE |
*Apoptosis is GENETICALLY ENCODED *Think of it as a "very dignified, neat, self-enclosed death" -- Cell does NOT pop open...membrane remains intact *It is a UBIQUITOUS PATHWAY *Enables cells to undergo HIGHLY REGULATED DEATH IN RESPONSE TO PRO-DEATH SIGNALING *Important in: - Normal digit development & development of brain & immune system - Normal maintenance of tissues & cell population *Without Apoptosis, normal development can't occur |
|
|
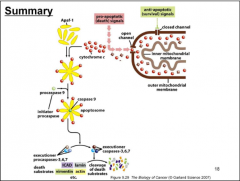
APOPTOSIS: MECHANISM'S 3 KEY EVENTS |
1. Initial Event - Cytochrome C's sudden release from mitochondria --> cytosol 2. Lipid Assymetry of membrane breaks down -- Phosphotidyl Serine (PS, shown in red fluorescent in video) becomes exposed on outside of cell -- labels dead cell to be rapidly consumed by macrophages 3. Although cell does NOT lyse...membrane becomes permeable to small molecules that can bind to DNA |
|
|
CORE APOPTOTIC SIGNALING COMPONENTS |
Pro-apoptotic protein ---> Anti-apoptotic protein (e.g. Bcl-2) --> Pro-apoptotic adaptor protein --> Cysteine protease --> APOPTOSIS for cells that are unhealthy/fated to die
*Pro-apoptotic Adaptor Protein - Like an "ON" switch *Anti-apoptotic Adaptor Protein - Inhibit the Pro-Apoptotic Adaptor Proteins in normally functioning cells that do not need to undergo Apoptosis --> Thus, Cysteine Proteases remain inactive --> Apoptosis is prevented *Cysteine Proteases - Once active, they begin cleaving proteins that regulate nuclear membrane integrity/cellular metabolism & DNA -- The "fuse is Lit" once these are activated |
|
|
BCL-2 |
*Bcl-2 is the founding member -- the classic anti-apoptotic protein
*In B-Cell Lymphoma, Bcl-2 is over expressed -- first disease it was observed in (hence the name)
*Bcl-2 activity -- depends on proper localization @ mitochondria --> Bcl-2 goes btwn cytosol & mitochondria, but for it to be truly active it needs to be on surface of mitochondria |
|
|
BAX & BID |
*BAX --> belongs to Bax Family of Proteins *BID --> belongs to BH3-only Family of Proteins *Both are Mammalian pro-apoptotic proteins
*Pro-apoptotic Protein expression often altered in cancer -- Often downregulated in cancer *BH3-only Pro-Apoptotic Proteins antagonize anti-apoptotic family members |
|
|
CASPASES |
*Caspases are Mammalian Cysteine Proteases *They are the central executioners of Apoptosis *Caspases "Karate chopping proteins" - Proteases responsible for the morphological & biochemical aspects of apoptosis - There are 14 caspases in Mammals *Cysteine Dependent Aspartate Directed Proteases *Exist as inactive zymogens activated by cleavage at aspartate residues within themselves * Have an ASPHOLE |
|
|
CLASSIFICATION OF CASPASES |
INITIATOR CASPASE - Caspases 8, 10, 9, 2 -Containing LONG N-terminal pro-domain -Auto-activated -DED: Caspase 8 and 10 contain a death- effector domain -CARD: Caspase 9 and 2 contain a caspase activation and recruitment domain (CARD)
EFFECTOR CASPASES - Caspases 3, 7, 6 -Containing SHORT N-terminal pro-domain -Activated by Initiator Caspases
|
|
|
APAF-1 |
*Type of Mammalian pro-apoptotic adapter protein *Activation of caspases --> Depends on Adapter Protein termed Apoptotic Protease Activating Factors |
|
|
ACTIVATION OF CASPASES |
*3 factors together (Apaf-1, ATP, Cytochrome C) comprise the apoptosome which activates initiator caspases
- Recall: Cytochrome C is a critical mobile carrier involved in ETC & ATP production, normally retained in the mitochondria....when it leaves the mitochondria, bad things happen...like the activation of Caspases |
|
|
HOW IS CYTOCHROME C RELEASE COORDINATED? |
BH3-only pro-apoptotic proteins play key role: *They inhibit anti-apoptotic proteins *AND activate multi-domain pro-apoptotic proteins *Multi-domain Pro-Apoptotic Proteins then form pores & release Cytochrome C
FYI: This is why over-expression of Bcl-2 blocks cell death |
|
|
HOW ARE BH3-ONLY PRO-APOPTOTIC FAMILY MEMBERS REGULATED SO THAT THEY CAN TIP THE BALANCE TOWARDS DEATH? |
BH3-only proteins are regulated by a variety of mechanisms including transcription and post-translational modifications |
|
|
SUMMARY OF CELL DEATH DIAGRAM |
|
|
|
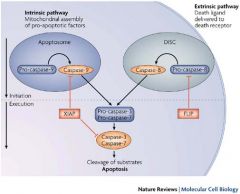
APOPTOSIS: INTRINSIC PATHWAY |
*The intrinsic, or mitochondrial, pathway is initiated from within the cell *Involves mitochondrial assembly of pro-apoptotic factors - Formation of Apoptosome *Works through CASPASE 9 *Then CASPASE 9 interacts with CASPASES 3 & 7 in the cleavage of proteins & execution of Apoptosis |
|
|
APOPTOSIS: EXTRINSIC PATHWAY |
A.k.a. Cell-Death Receptor Pathway
*Extrinsic Pathway involves delivery of death ligand to death receptor
*Begins outside the cell via activation of pro-apoptotic receptors on the cell surface (TNF Receptor, FasL Receptor) -- aka the DEATH RECEPTORS - TNF: Tumor Necrosis Family - Characterized by extracellular Cysteine Rich Domains (CRDs) *Involves formation of DISC *This pathway signals thru Adaptor Protein to activate CASPASE 8 *Then CASPASE 8 interacts with CASPASES 3 & 7 in the cleavage of proteins & execution of Apoptosis
|
|
|
INTRINSIC VS. EXTRINSIC PATHWAYS COMPARED |
|
|
|
NECROSIS |
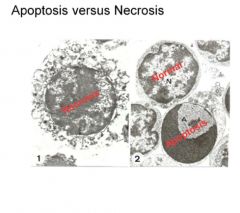
Necrosis is diff. from apoptosis in that it is NOT programmed and is "messy"
*Disordered, not programmed *No signaling *Cell contents spill out *Promotes tissue injury & inflammation
|
|
|
APOPTOSIS VS. NECROSIS SIDE BY SIDE COMPARISON |

|
|
|
Where is erythropoietin produced? |
Kidneys |
|
|
What is Erythropoietin produced in response to? |
Lack of oxygen |
|
|
How long does erythropoesis take? |
7 days |
|
|
Why is the red blood cell's life span only 120 days? |
The components of the cell wear out, and without a nucleus, they are unable to replace them |
|
|
What is the incidence of sickle cell anemia? |
1:500 African American children |
|
|
True or False: "The longer a cell spends in G1, the more likely it is to give rise to differentiated daughter cells." |
True |
|
|
What causes Sickle Cell Anemia? |
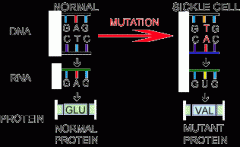
Mutation of a single nucleotide (SNP) from A-->T on the beta globin gene.
Results in Glutamic Acid --> Valine substitution at position 6
|
|
|
THE CENTRAL DOGMA |
1. DNA can replicate itself ---> REPLICATION |
|
|
GOAL OF DNA REPLICATION |
To replicate genomic contents of a single cell in order to allow # of cell divisions it takes to form an organism
(which is about 7*10^13 divisions) |
|
|
5 CHALLENGES ASSOCIATED WITH DNA REPLICATION |
1*Must reproduce 6.4 * 10^9 base pairs in the 4-6 hours that cell division allows...which is roughly size of 2 DVD's ~300,000 bp/s so must be highly efficent
2*Must be of high fidelity to avoid errors (Error rate = 1/10^9 or 1/10^10 bp incorporated)
3*DNA is highly coiled in the nucleus & associated w/proteins to form chromatin --> largely inaccessible to replication enzymes
4*Polymerases necessary for DNA replication only work in 1 direction but DNA replication occurs in 2 --> Challenge is replicating both DNA strands simultaneously
5*DNA replication has to be limited to once/cycle --> Challenge is stopping replication after it has been initiated |
|
|
MECHANISM OF DNA REPLICATION (Think in terms of conservation) |
*DNA replication is SEMI CONSERVATIVE This means there's a parental strand and a new daughter strand after replication
*Uses Watson-Crick base pairing to duplicate DNA |
|
|
ORIGIN OF REPLICATION |
Site within DNA where replication is initiated
Other Things to Know about Origin: 1. Characterized by specific DNA sequences recognized by group of proteins called Origin Recognition Complex (ORC) 2. ORC leads to further protein recruitment to assemble pre-replication (pre-RC) 3. In humans, DNA recognition sequence can vary widely 4. E.coli has a single replication origin (replicon) while humans have up to 100,000 replication origins in a single cell 5. Replication origins are typically A-T rich because easier to separate than G-C due to having 2H bonds instead of 3
|
|
|
WHAT IS A REPLICON? |
A replicon is a DNA or RNA molecule, or a region of DNA or RNA, that replicates from a single origin of replication.
*For most prokaryotic chromosomes, the replicon is the entire chromosome
*For eukaryotic chromosomes, there are multiple replicons per chromosome |
|
|
THE REPLICATION FORK |
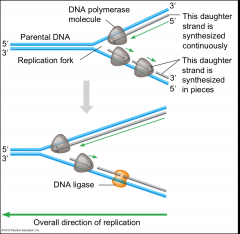
Forms when DNA strands have been sufficiently unwound to allow replication to take place
*Located AHEAD of parental DNA, and BEHIND replicated DNA |
|
|
STAGES OF DNA REPLICATION |
1. Unwinding DNA to form replication fork
2. Stabilization of single stranded DNA
3. Initiation/Priming
4. Elongation (includes proofreading & editing) of the Leading Strand (5' --> 3')
5. Lagging strand synthesis (3' --> 5') -- it is discontinuous
6. Licensing: Ensuring that each replicon only replicates once/cycle |
|
|
HOW REPLICATION FORK IS FORMED |
- DNA strands separated in ATP-dependent fashion by helicases
- They bind to single strand & move thru DNA like a zipper |
|
|
PROBLEMS WITH DNA'S INTRINSIC TWIST |
DNA has an intrinsic twist (360 degrees for every 10bp) but is overwound (supercoiled) in order to facilitate packaging in nucleus --> creates problems when strands are separated by helicase
|
|
|
ROLE OF TOPOISOMERASES |
DNA becomes overwound ahead of the replication fork. Its supercoils can delay/interfere w/advance of replication fork
This DNA Twist problem is alleviated by topoisomerase enzymes - they relax the supercoiling in DNA by making breaks/nicks that lessen tension
Topoisomerases are often indicated in cancer |
|
|
WHY DOES SINGLE STRANDED DNA (ssDNA) NEED TO BE STABILIZED & HOW IS THAT ACHIEVED? |
*Newly separated ssDNA will try & satisfy unpaired bases - ends up forming hairpin helices that restrict passage of DNA polymerase
*SSBP = Single Stranded Binding Proteins - Stabilize newly formed ssDNA w/o covering the bases |
|
|
CHARACTERISTICS OF DNA POLYMERASES |
1. Enzymes that catalyze polymerization of deoxyribonucleotides into a DNA strand (like a giant hand that wraps around DNA as it moves thru the hand) 2. Magnesium ion used for catalytic activity 3. Can only add free nucleotides to 3' end 4. Cannot form new strands 5. Synthesize DNA only in 5' --> 3' direction (required for accuracy) 6. Misincorporation rate = 1*10^6 (Note: Lower than actual rate of fidelity of DNA replication) 7. Have proofreading ability (exonuclease activity that removes errant base pairs), therefore bringing the fidelity rate up to 1/10^9 or 1/10^10) |
|
|
3 MAJOR POLYMERASES ASSOCIATED W/DNA REPLICATION (IN HUMANS)
*There are 15 total but 3 major ones |
1. Polymerase alpha = RNA PRIMASE 2. Polymerase delta = Lagging strand synthesis 3. Polymerase epsilon = Leading strand synthesis
|
|
|
POLYMERASE ALPHA DETAILS |
(aka RNA PRIMASE)
-Synthesizes an RNA Primer (removed later on) -Then acts as DNA pol to elongate strand for approx 20 bp before delta & epsilon take over
*UNLIKE OTHER DNA POLYMERASES, POLYMERASE ALPHA CAN INITIATE A NEW STRAND
|
|
|
POLYMERASE DELTA DETAILS |
LAGGING STRAND SYNTHESIS
-Highly processive (means it can synthesize a lot) -Proofreads 3' --> 5' -Exonuclease activity
|
|
|
POLYMERASE EPSILON DETAILS |
LEADING STRAND SYNTHESIS
-Highly processive -Proofreads 3' --> 5' -Exonuclease activity -Related to polymerase delta |
|
|
MAJOR POLYMERASES IN PROKARYOTES
*There are 5 major ones, but 3 we need to know |
1. DNA POL I -- DNA repair & removal of primers 2. DNA POL II -- DNA repair 3. DNA POL III -- Main polymerase in bacteria |
|
|
PROCESSIVITY OF POLYMERASE DELTA & EPSILON |
- Polymerases fall off DNA strand, thereby curtailing their ability to extend strands
- To extend processivity, Polymerase Delta and Polymerase Epsilon associate with protein PCNA ("clamp proteins")
*FYI: PCNA = Proliferating Cell Nuclear Antigen |
|
|
THE LAGGING STRAND ISSUE & SEMI DISCONTINUOUS SYNTHESIS |
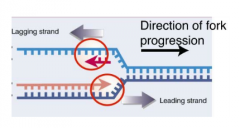
DNA polymerases extend DNA template ONLY in 5' --> 3' direction
This is OK for one DNA strand but not the other, given that the replication fork moves in 1 direction. That is why you end up with Semi Discontinuous Synthesis |
|
|
HOW SEMI-DISCONTINUOUS SYNTHESIS IS ACHIEVED |
1. Priming of leading strand 2. Initiation of leading strand synthesis 3. Priming of lagging strand 4. Synthesis of first Okazaki fragments 5. Priming/Synth. of second Okazaki fragments |
|
|
CLEAN UP PROCESS ON LAGGING STRAND |
-RNA Primers have to be removed before the polymerase reaches it (i.e. before next round of synthesis)
-DNA Polymerases repair/fill in missing segment
-DNA Ligases join Okazaki fragment to new strand (seal together) |
|
|
THE REPLISOME |
The replisome is a complex molecular machine that carries out replication of DNA.
The replisome is composed of a number of proteins including helicase, PCNA, topoisomerase, primase, DNA polymerase I, and ligase.
This complex moves along dsDNA like zip of a zipper
**To facilitate this, lagging strand must bend back |
|
|
KEY POINTS OF DNA REPLICATION |
1. DNA Replication is a HIGH FIDELITY, HIGHLY EFFICIENT process that allows complete duplication of human genome in matter of hours
2. Major characteristic of DNA Replication is the REPLICATION FORK, which can arise on up to 100,000 origins within the cell
3. A lot of different proteins are involved in DNA replication -- they form a "machine" known as the REPLISOME to facilitate replication
4. Replication is asymmetric, w leading & lagging strands that need to be coordinated
5. Replication occurs in a # of STAGES: (1) Unwinding (2) Strand separation (3) Priming (4) Elongation (5) Clean Up
6. Replication coordinated w/cell cycle --> LICENSING -- this ensures duplication of DNA content takes place only once per cell division |
|
|
DNA REPLICATION IN THE CELL CYCLE (LICENSING) |
- DNA replication in cell is confined to 4-6 hrs during "S Phase"
- 100,000 replicons replicate DNA at this time, but each cannot replicate more than once due to Licensing of DNA Replication Origins
- Licensing helps prevent GENETIC INSTABILITY |
|
|
GENETIC INSTABILITY |
Uneven distribution of genes; It is a hallmark of cancer |
|
|
HOW IS LICENSING ACHIEVED? |
- ORC proteins bind to Origin throughout cell cycle - Cdc6 is recruited. Mcm proteins (i.e. helicase) recruited too. They form pre-RC. -CDK activates pre-RC to initiate bidirectional replication forks -Cdc6 destroyed upon initiation until next cycle. No new initiation can occur without it - thus no more than 1 replication/cycle |
|
|
WHAT ABERRANT CONDITIONS ILLUSTRATE NEED FOR CONTROLLED, HIGH FIDELITY DNA REPLICATION? |
1. Genetic disorders (e.g. Ataxia Telangiectasia) - decreased fidelity results in increased mutation rate
2. Uncontrolled proliferation (e.g. Cancer, Autoimmune Disorders- can also be promutagenic)
3. Bacterial & Viral Replication
THESE DISEASES CAN BE TARGETED W/DRUGS THAT AFFECT DNA REPLICATION (FOR THERAPEUTIC PURPOSES) |
|
|
EXAMPLES OF DRUG CLASSES THAT TARGET DNA REPLICATION FOR THERAPEUTIC PURPOSES |
1. DNA Damaging Agents -- i.e. Cisplatin, popular in treatment of cancers
2. Topoisomerase Inhibitors -- if you interfere w/topoisomerase inhibitors, DNA will be under stress & replication stops
3. Anti-Metabolites -- we can design small molecules that will fit into the ATP pocket of the enzyme and interfere w/its activity
4. Nucleoside Analogs - main drug used in HIV is a nucleoside analog (derivative of one of the 4 bases) that prevents RNA synthesis from proceeding forward |
|
|
OKAZAKI FRAGMENTS |
Short lengths of DNA synthesized btwn RNA primers on the lagging strand of the replication fork |
|
|
4 IMPORTANT CONDITIONS FOR A CELL TO PROLIFERATE CORRECTLY |
- correct & accurate duplication of genome
- correct & accurate increase in organelles
- sufficient nutrients & metabolites for replication
- mitotic machinery functioning correctly |
|
|
CELL GROWTH & DIVISION - WHAT'S THE DIFFERENCE? |
Growth w/no cell division --> ONE BIG CELL
Cell division w/no growth --> BUNCH OF TINY LITTLE CELLS (molecule involved is WEE1 - mutation would make cells shrink)
Cell division + Growth = PROLIFERATION, YAY! |
|
|
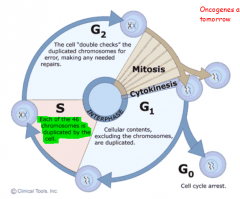
THE CELL CYCLE DEFINED |
Eukaryotic cell cycle is orderly series of events whereby a single cell replicates its contents & divides into 2 cells. Cell Cycle can be seen as master regulator of DNA synthesis
Consists of these stages:
INTERPHASE G1: Cellular contents, EXCLUDING chromosomes, are duplicated S: Each of the 46 chromosomes is duplicated G2: Cell double checks duplicated chromosomes for errors, making any needed repairs MITOSIS M: Mitosis CYTOKINESIS Cytoplasmic Division
|
|
|
G0 (G Zero Phase) |
G0 = Quiescent state; Resting phase
Many mammal cells, such as the neuron, remain permanently or semipermanently in G0.
Some cells enter G0 temporarily & divide infrequently, like hepatocytes (liver cells) |
|
|
Regulatory Checkpoints in Cell Cycle |
*The regulatory checkpoints lie at or near the points of transition within interphase
*The 1st checkpoint is located at the end of the cell cycle's G1 phase, just before entry into S phase, making the key decision of whether the cell should divide, delay division, or enter a resting stage
*The 2nd checkpoint is located at the end of G2 phase, triggering the start of the M phase (mitotic phase). In order for this checkpoint to be passed, the cell has to check a number of factors, such as DNA damage via radiation, to ensure the cell is ready for mitosis. |
|
|
DNA CONTENT THROUGHOUT CELL CYCLE (for animal cells) |
*n = haploid *2n = diploid
G1: 2n S: Cells go from 2n --> 4n G2: 4n M: 4n After Cytokinesis: 2n |
|
|
WHY WOULD A LIVER CELL WANT TO DIVIDE INFREQUENTLY & ENTER G0? |
-Hepatocytes exposed to lots of toxins
-Temporary exit into G0 limits the pool of regeneration going on in the liver by controlling the proliferation of hepatocytes |
|
|
CYCLIN-DEPENDENT KINASES (CDK'S) - WHAT ARE THEY? |
-Family of small protein kinases (~30kDa) -Regulate large # of cellular functions including transcription/mRNA processing/cellular differentiation -Level of CDK's remains fairly constant, but their activity during cell cycle changes, depending on level of activators present (activators are known as cyclins) |
|
|
KINASES |
Carry out phosphorylation, use ATP
"KINdly put a phosphate on" |
|
|
CYCLINS |
Family of proteins that form complexes w/cdk's to yield an active kinase
Cyclin expression levels are regulated by expression of the previous Cyclin -In other words...Cyclin's interaction w/CDK control the synthesis of subsequent Cyclin/CDK's
|
|
|
CYCLINS & CDK'S AND THE MATCHING CELL CYCLE PHASE |
G1 -Cyclin D with CDK 4, CDK 6
G1/S -Cyclin E with CDK 2
S -Cyclin A with CDK 2 (again)
M -Cyclin B with CDK 1
Mnemonic/Peg List: *Brad Pitt's Dick is 4-6 inches long --> G1, Cyclin D, CDK 4/6 *Brad Pitt's Sh*t is the size of 2 Elephants --> G1/S, Cyclin E, CDK 2, *Sh*tting with 2 A*ssholes is hard --> S, Cyclin A, CDK 2 *1 Motherf*cking B*tch --> M, Cyclin B, CDK 1
|
|
|
D-TYPE CYCLINS |
There are 3 in mammals: -D1, D2, D3 |
|
|
Maturation Promotion Factor (MPF) |
Cyclins w/respective CDK form the MPF
Cyclins can be divided into 4 basic categories based on their period of expression during cell cycle (i.e. M-Cdk is trigger mitosis machinery and S-Cdk is trigger DNA replication machinery)
Progression thru each phase of cell cycle is combined efforts of cyclin (Regulatory subunit) and CDK's (Catalytic Subunit) |
|
|
Important checkpoints in cell cycle |
1. G1/S Checkpoint
2. G2/M Checkpoint
3. Spindle Checkpoint |
|
|
CDK INHIBITORS |
-Cyclin-dependent kinase inhibitor - proteins that perturb cyclin/CDK interaction
-They slow down or arrest progression of the cell cycle through this interference
--> Often important in cell cycle checkpoints --> Referred to as Tumor Suppressor Proteins (bc mutations of these proteins associated w/tumors) --> Nomenclature derives from their motility on SDS-PAGE (gel-electrophoresis)
e.g. p16, p53
|
|
|
p16 |
-Important in transition from G1/S
-Frequently mutated in cancers
-Associated w/prevention of melanoma & esophageal cancer |
|
|
p53 |
"GUARDIAN OF THE GENOME"
-p53 mutated/missing in 50% of tumors
Serves 3 Major Purposes: (1) Activate DNA repair mechanisms (2) Arrest cell cycle at G1/S checkpoint (3) Induce Apoptosis in cells where DNA damage wasn't corrected
-basic functions of p53 regulated thru interactions with mdm2....via UBIQUITINATION
-p53 comes into play in 3 instances: 1. DNA Damage 2. Cell cycle abnormalities 3. Hypoxia
-DNA Damage activates cell cycle checkpoints via the activity of p53 |
|
|
Hypoxia |
Condition in which the body or a region of the body is deprived of adequate oxygen supply |
|
|
WAYS IN WHICH DNA DAMAGE CAN OCCUR |
-Ionizing Radiation
-UV light
-Oxidation from metabolites
*Damage can vary from 10^4 to 10^6 lesions per cell per day |
|
|
3 POSSIBLE FATES FOR CELL W/SIGNIFICANT DNA DAMAGE |
1. Senescence - irreversible dormancy (cell won't be able to divide again, and may lose most/all of its functionality too; not the same as G0)
2. Apoptosis - programmed cell death
3. Cancer |
|
|
WHAT DETECTS DNA DAMAGE/ACTIVATES THE CHECKPOINTS? |
*Two serine/threonine kinases are instrumental in detecting DNA Damage
*If DNA Damage is detected, cell cycle checkpoint is activated via activity of p53 |
|
|
ATAXIA TELANGIECTASIA (ATM) |
Kinase protein mutated in AT --> responds to double strand breaks & chromatin disruption and works thru activity of p53 --> p21
Disease associated w/cerebellar degeneration (causes pt to shiver/shake)
|
|
|
ATAXIA TELANGIECTASIA & RAD3 RELATED PROTEIN (ATR) |
Kinase that get's activated by stalled replication forks
Also works thru activity of p53 --> p21 |
|
|
p53 & DNA damage response for G1 arrest |
-DNA damage activates ATM/ATR pathways -Leads to activation of p53 -p53 transactivates p21 Wafl/Cip1 (a CDK inhibitor) -Results in G1 arrest
*Targeted disruption of p21 Wafl/Cip1 gene shown to compromise G1/S checkpoint |
|
|
p53 & G2/M arrest |
-Progression of cells from G2 to Mitosis is driven by Cyclin B and CDK 1
-p53 shown to induce G2/M arrest by inhibiting activation of cyclin B/CDK1, again thru p21
-Cyclin B/CDK1 activation prevented by repressing cdc25C --- on account of phosphorylation by Chk1 |
|
|
WHAT IS A TATA BOX? |
TATA BOX:
-A DNA sequence recognized by components of the General Transcription Factors (most notably, site at which RNA polymerase binds DNA)
-Located within promoter, upstream of transcription site |
|
|
WHERE IS THE TATA BOX LOCATED? |
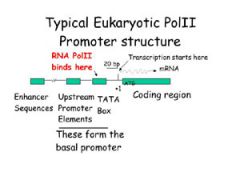
LOCATED WITHIN PROMOTER, UPSTREAM OF TRANSCRIPTION SITE
(at about -25) |
|
|
What is Gene Regulation? |
Fundamental process by which gene pdts (RNA, and finally protein) are controlled
*Remember - All cells have the SAME DNA...what defines their identity is the 'suite of genes' that are active vs. inactive |
|
|
Why is Gene Regulation important? (i.e. What processes depend on Gene Regulation?) |
Turning ON & OFF of genes (or up & down of genes) is the goal of many pathways.
Gene Regulation is Key to: 1. Embryology & Development (regulation of proliferation/differntiation) 2. Terminal cell differentiation (Identity) & therefore function 3. Cellular response to disease & injury |
|
|
What is the pdt of transcription? |
RNA, ultimately.
-But 1st, DNA transcribed into Pre(hn)-mRNA -Then RNA processing (splicing) takes place to create mRNA -Then mRNA leaves the Nucleus & goes to Cytoplasm (Nuclear export) for translation
|
|
|
GENE EXPRESSION ON THE TRANSCRIPTIONAL SIDE IS INVOLVED IN THE REGULATION OF...? |
RNA levels in the cell
Gene Regulation is able to regulate RNA levels by: 1. Controlling access to basic resources (i.e. DNA as a template) 2. Production of the product from basic resources 3. Regulating the lifetime of the pdt |
|
|
What does mRNA concentration (in the cell) reflect? |
Balance btwn Synthesis & Degradation |
|
|
At what point can RNA be degraded? |
After transcription, when it leaves nucleus --> cytoplasm via Nuclear Export
That way it does not get translated into a protein |
|
|
STAGES OF TRANSCRIPTIONAL GENE REGULATION |
1. Gene Activator protein binds to chromatin 2. Chromatin Remodeling (with Chromatin Remodeling Complex) 3. Covalent Histone Modification (with Histone Modification Enzymes) 4. Additional Activator Proteins bind to gene regulatory region 5. Assembly of pre-initiation Complex at the Promoter (once general TF & RNA pol come) 6. Transcription Initiation |
|
|
WHAT IS TRANSCRIPTIONAL SYNERGY? |
Gene Activators & Gene Repressors - their combined effect on Transcription exceeds the sum of the individual effects |
|
|
What % (approximately) of human proteins are involved in regulation of gene expression? |
5-10% |
|
|
Levels of Chromatin Organization |
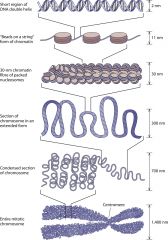
1. DNA wraps around histone proteins, forming nucleosomes 2. The nucleosomes arranged like "beads on a string" are Euchromatin (genes under *Active* transcription) 3. DNA coding *Inactive* genes ("turned off" or "silent") are more tightly packaged as Heterochromatin
|
|
|
HOW IS CHROMATIN'S OVERALL STRUCTURE AFFECTED BY CELL CYCLE PHASE? |
Chromatin's overall structure depends on the stage of the cell cycle: -During interphase, chromatin is structurally loose to allow access to RNA and DNA polymerases that transcribe and replicate the DNA. -In & just before entering mitosis/meiosis, the chromatin packages more tightly to facilitate segregation of the chromosomes during anaphase
|
|
|
HOW IS CHROMATIN'S LOCAL STRUCTURE AFFECTED BY GENE EXPRESSION? |
The local structure of chromatin depends on the genes present on the DNA:
*DNA coding active genes are actively transcribed ("turned on") & more loosely packaged (euchromatin)
*DNA coding inactive genes ("turned off") are found associated with structural proteins and are more condensed/tightly packaged (heterochromatin) |
|
|
What are the 2 major mechanisms used in Chromatin Remodeling? |
1. COVALENT HISTONE MODIFICATION
2. ATP-DEPENDENT CHROMATIN REMODELING (i.e. SWI/SNF) |
|
|
WHAT ARE COVALENT HISTONE MODIFICATIONS? |
Acetylation, methylation*, phosphorylation, and ubiquitination:
These processes affect packing of chromatin & affinity of DNA for protein.
*this methylation is different from DNA methylation
Mnemonic: Histone Acetylation makes DNA Active (b/c it relaxes DNA coiling, allowing for transcription)
|
|
|
WHAT DOES ATP-DEPENDENT CHROMATIN REMODELING REFER TO? |
Move, eject, or restructuring of nucleosomes |
|
|
WHAT ROLE DOES ACETYLATION PLAY IN CHROMATIN REMODELING? |
*Acetylation = transfer of an acetyl group
*It is a type of Covalent Histone Modification
Acetylation removes the (+) charge on the histones, thereby decreasing the interaction of the N termini of histones w/negatively charged phosphate groups of DNA.
As a consequence, the condensed chromatin is transformed into a more relaxed structure that is associated with greater levels of gene transcription.
|
|
|
WHAT IS THE KEY ENZYME IN ACETYLATION? |
Acetyl-Coenzyme A |
|
|
WHAT AMINO ACID RESIDUE ON THE N-TERMINUS OF HISTONES IS ACETYLATED DURING ACETYLATION? |
LYSINE
Recall: Lysine is a + charged amino acid (a base) |
|
|
WHAT ROLE DOES METHYLATION PLAY IN CHROMATIN REMODELING? |
Methylation of histones does the OPPOSITE of Acetylation
*Histones can be methylated on lysine (K) & arginine (R) residues but methylation is most commonly observed on lysine residues of histone tails
Mnemonic: Histone Methylation Mostly Makes DNA Mute (bc usually represses DNA transcription - tho not always) |
|
|
WHAT IS EPIGENETIC REGULATION OF GENE EXPRESSION? |
Gene silencing - prevents expression of a certain gene; can occur either during Transcription or Translation
Mechanisms of Epigenetic Silencing: Chromatin remodeling (i.e. histone deacetylation, histone methylation) and DNA methylation |
|
|
WHAT ARE SOME EXAMPLES OF THE MECHANISMS OF EPIGENETIC SILENCING (TRANSCRIPTIONAL)? |
*Chromatin Remodeling - specifically Histone deacetylation and Histone methylation
*DNA Methylation |
|
|
WHAT IS DNA METHYLATION?
|
Process where a methyl group is added to the cytosine or adenine DNA nucleotides
(NOTE: DIFFERENT FROM HISTONE METHYLATION) |
|
|
WHAT IS CONSTITUTIVE TRANSCRIPTION? |
Constitutive Transcription - when genes are transcribed (and thus expressed) all the time (i.e. for glyclosis & housekeeping proteins)
UNLIKE regulated transcription |
|
|
WHAT IS REGULATED TRANSCRIPTION? |
When rate of gene transcription is controlled, for example, by helping or hindering RNA polymerase binding to DNA
-Exists b/c some proteins only need to be produced at specific times or by certain cells. (Don't need to be constitutively expressed)
-Regulation of transcription carried out by transcription factors
|
|
|
What are Transcription Factors? |
*TRANSCRIPTION FACTORS: sequence-specific DNA-binding proteins that function in controlling the rate of transcription
*Make contact w/sides of the bases exposed in major groove of B-DNA (bc that is where they base edges are most accessible)
*Key feature of Transcription Factors is that they have DNA Binding Domain(s) which are often further characterized by a recognition helix |
|
|
What are Enhancers? |
Regions of DNA that alters gene expression thru the binding factors of Transcription Factors
*Opposite of Silencers, basically
|
|
|
What are Silencers? |
Region of DNA where negative repressors bind |
|
|
Where are Enhancers/Silencers located on the eukaryotic gene? |
Enhancers & Silencers may be located close to, far from, or within the introns of the gene whose expression it regulates. These regions may be 1000's of bp away from promoter region!
*But in lecture/diagrams, most often we see the Enhancers upstream of the transcription start site |
|
|
How do Transcription Factors contribute to transcription regulation? |
Function in controlling the rate of transcription
*Often, they activate transcription *But they ONLY interact w/specific genes (that's why most eukaryotes have > 1,000 TF's) *Transcription Factors most often modified/activated via phosphorylation *Activated Transcription Factors bind to Enhancer Regions, often upstream of transcription start site. Once bound to DNA, the TF will move towards transcription start site to interact with Transcription Complex. *Once formation of Transcription Complex is complete, transcription can begin |
|
|
6 TYPES (OR EXAMPLES) OF DNA BINDING DOMAINS |
*Their names describe their structure* 1. Helix-turn-helix: repressor proteins, regulators of developmental processes) 2. Zinc Finger: Zn ion stabilizes finger-like structure (specific triplet of base pairs) 3. Leucine Zipper: Leucine every 7th residue; Zips up to dimeriZe proteins 4. Winged Helix: 4 helices & 2-strand beta sheet 5. Winged Helix turn Helix: 3-helical bundle & 4 strand beta sheet 6. Helix loop Helix: Two helices connected by a loop (associated w/ some transcription factors) |
|
|
WHAT DO COACTIVATORS DO? |
Because enhancer/repressor regions are sometimes located far from the Promoter, a protein that rearranges DNA's structure to bring them closer is sometimes needed -- thats what Coactivators do!
Coactivators bind to Activators or Transcription Factors, not to DNA directly (?) |
|
|
TRANSCRIPTIONAL CONTROL IN PROKARYOTES |
Transcription is where most regulation occurs in prokaryotes b/c translation is coupled.
Classic example is Lac Operon |
|
|
LAC OPERON - WHAT IS IT AND WHAT DOES IT SIGNIFY? |
-Consists of 3 genes needed for digestion of lactose (to be utilized for NRG in absence of glucose)
-Represents the idea of an OPERATOR region that binds to a REPRESSOR: A tetrameric Repressor protein (I gene) is localized to Operator to prevent RNA pol read-thru
-In absence of glucose & presence of lactose, Allolactose will bind to repressor subunit, inducing a conformational change that reduces the repressor's affinity for the lac operon. cAMP (signals starvation) forms CAP-cAMP, which promotes RNA pol attachment.
-In presence of glucose, no CAP-cAMP forms & poor read-thru of RNA pol -- thus, digestion of lactose will not occur
This makes sense b/c Glucose is preferred as an NRG source over Lactose |
|
|
WHAT IS A DNA MICROARRAY & WHAT DOES IT HELP DIAGNOSE? |
- Collection of microscopic DNA spots attached to a solid surface (looks like a chip) - Measures expression levels of large # of genes simultaneously -Useful in diagnosing cancer - for example if you were to hybridize the spots w/mRNA's from Head & Neck Cancers |
|
|
THE VARIOUS TYPES OF RNA'S & THEIR CLASSIFICATION |
1. Coding RNA - mRNA
2. Non-coding RNA = ncRNA - Transcription RNAs (rRNA & tRNA) - Small RNAs (such as miRNA, siRNA, piRNA, snoRNA, snRNA) - Long non-coding RNA (such as XIST and HOTAIR)
|
|
|
What is miRNA? |
MicroRNA (22-24bp in length) - involved in Translational control, and can work as both Positive & Negative regulators of cellular function; Can control transcript degradation, translation repression, potentially transcriptional repression as well. Very old.
-Human genome encodes > 1,000 miRNA's that target ~60% of our genes! Different sets for different tissues
-Type of non-coding, small RNA |
|
|
What is siRNA? |
Small interfering RNA (22bp in length) - RNA interference
-Useful in treating Choroidal neovascularization (CNV); Vascular Endothelial Growth Factor (VEGF) and its receptor are targeted using siRNA. These molecules control growth of new blood cells.
-Type of non-coding, small RNA |
|
|
What is Choroidal Neovascularization (CNV)? |
Choroidal neovascularization (CNV), an age-related growth of new blood vessels in retina that leads to blindness
It is a common symptom of macular degeneration
|
|
|
What is piRNA? |
PIWI-interacting RNA - Transposon silencing
-Type of non-coding, small RNA |
|
|
What is XIST? |
Xist - On the X chromosome; acts as major effector of the X inactivation process
-Type of non-coding, long RNA |
|
|
What is HOTAIR? |
HOX Antisense Intergenic RNA - faculty silencing
-Type of non-coding, long RNA |
|
|
Problems w/using siRNA in Therapeutics |
1. Ensuring good, dose-dependent delivery to targets 2. Off-target effects |
|
|
How Does Genetic Information Get Amplified to Organismal Phenotype? |
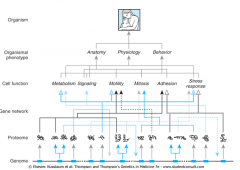
Genome structure and corresponding pathways of “amplification” of genetic information facilitate multi-level expansion of the information available to influence and regulate Cellular Function & Organismal Phenotype. |
|
|
KEY STRUCTURAL FEATURES OF A HUMAN GENE |
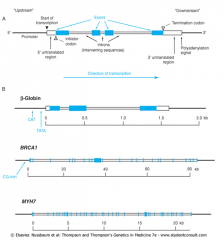
Typical human gene has Exons, Introns, and a Promoter |
|
|
β-globin (Beta-globin) |
Very small gene (only 3 exons) involved in hemoglobin disorders - (e.g. Beta-thallassemia, Sickle Cell Disease) |
|
|
BRCA 1 |
Gene that has 24 exons
Associated w/inherited BREAST and OVARIAN CANCER susceptibility |
|
|
Beta-myosin heavy chain (MYH7) |
Large gene (40 exons)
Mutations on this gene associated w/inherited Hypertrophic Cardiomyopathy |
|
|
KEY CHARACTERISTIC OF GENETIC CODE |
The genetic code is DEGENERATE. Because there are 61* possible combinations for triplets of nucleotides (i.e. codons), an Amino Acid can have multiple corresponding codons.
*64 – 3 (for the STOP codons which don’t code for an Amino Acid) = 61 triplets |
|
|
START CODON & MATCHING AMINO ACID |
AUG
- Encodes for Methionine (Met) when located in KOZAK SEQUENCE (GCCPuCCAUGG) |
|
|
STOP CODONS |
Mnemonic:
UGA = U Go Away
UAA = U Are Away
UAG = U Are Gone |
|
|
CODON USAGE IN NUCLEAR VS. MITOCHONDRIAL GENES – THE SAME OR DIFFERENT? |
Different! |
|
|
Significance of GT & AG sequences |
-Sequences at Exon/Intron borders -Associated w/RNA splicing
*GT & AG are SPLICE SITES* |
|
|
Polyadenylation Signal |
“AATAAA”
Important signal for addition of polyA tail, which protects mRNA from degradation |
|
|
Length of Poly A tail is significant because… |
Determines half-life of mRNA |
|
|
What is a CAAT Box? |
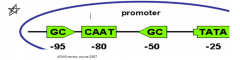
- Binds transcription factors to regulate amount of transcription - Located in Promoter at ~ -80
|
|
|
TATA Box binds to what? |
TFIID – Transcription Factor II D
*Determines the exact start site (not in all promoters) |
|
|
What is the Initiator (INR)? |
INR compliments the TATA Box to localize the transcription start site |
|
|
Challenge with large genes (such as Dystrophin) |
The larger the gene (in terms of kb), the more difficult it is to screen for
e.g. Dystrophin, associated w/muscular dystrophy, is one of the longest and most difficult ones (~2500 kb) |
|
|
Pseudogenes |
*Pseudogenes are Non-functional copies of a gene
- Usually not expressed - Often occur in gene families such as α and β-globin genes - Cause misalignment via erroneous chromosomal pairing during meiosis |
|
|
α-Thalassemia - What is it & what causes it? |
What is it? Blood disorder that reduces production of hemoglobin
What causes it? Deletion of 1 or more alpha-globin genes on Chromosome 16 which is the result of....
Misalignment of homologous pair & recombination btwn the α1 and α2 gene results in deletion/duplication of α-globin genes
*Illustrates effects of Unequal Crossing Over *Most common in Southeast Asians
|
|
|
What is the result of Unequal Crossing Over? |
If homologous chromosomes misalign during crossover…. · One pdt will contain an insertion · And the other pdt will have a deletion |
|
|
How are Clustered Human Repetitive DNA Sequences arranged? |
-As arrays of short repeats organized tandemly in Head-to-Tail Configuration
-Clustered Human Repetitive DNA Sequences comprise ~10-15% of human genome (evolutionary remnants) |
|
|
What does “satellite DNA” refer to? |
The tandem repeats of Clustered Human Repetitive DNA Sequences, collectively |
|
|
Length of the Tandem Repeat Arrays |
*Variable both in terms of total length, and the nucleotide repeat that comprises the array |
|
|
Applications of Tandem Repeats? |
· Routinely used in designing tools for Molecular Pathology · Also used in forensics, paternity testing, etc |
|
|
What is noteworthy about LONGER tandem repeats (e.g. α satellite DNA)? |
· Found at centromeres of human chromosomes · Important for attachment of chromosome to the microtubules of the spindle during cell division |
|
|
What are SINEs? |
SINEs = Short Interspersed Nuclear Elements · Include Alu sequences that are ~300bp · Make up ~10% of DNA |
|
|
What are LINEs? |
LINE = Long Interspersed Nuclear Elements · L1 Family · Up to 6 kb long · Accounts for ~20% of human DNA |
|
|
Familial Hypercholesterolemia (FH) is associated with what mutation(s)? |
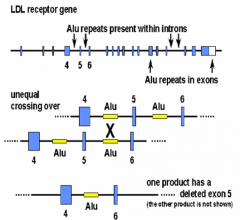
*Unequal Crossing Over with ALU repeats*
-Result: 1 pdt with a deleted exon & other product has an extra exon
- Affects the LDL (Low-Density Lipoprotein) Receptor gene, which leads to FH
|
|
|
What types of mutations are Alu & LINE Sequences associated with? |
· Insertional Activation: Some Alu & LINE generate copies of themselves & integrate at other genomic sites · Recombination & Unequal Crossing Over: Causes disease
|
|
|
What is the size of mitochondrial (mt) genome? |
37 genes (16,569 bp) |
|
|
Key Features of the Mitochondrial (mt) Genome & Structure |
· Closed, circular · NO INTRONS - all coding sequences are contiguous · Located in Mitochondrial Matrix · Exist in thousands of copies per cell · Has 2 strands: (1) a GUANINE-RICH heavy strand (2) a CYTOSINE-RICH light strand · Contains 13 polypeptide subunits, 22 tRNA genes and 2 rRNA genes
|
|
|
Issues with mitochondrial genome are typically characterized by... |
Fatigue & muscle weakness
Recall: Where do we have a high amount of mitochondria? MUSCLE! |
|
|
FEATURES OF MITOCHONDRIAL GENETICS |
*Maternal Inheritance *Mutation rate is higher than Nuclear DNA's *Replicative Segregation - No segregation btwn meiosis & mitosis *Mitochondrial DNA replicates by increasing # of genomes in proportion to mitochondrial mass; Does NOT ensure each genome replicates same # of times; Can lead to changes in representation of alleles in daughter mitochondria *Heteroplasmy - Mixed population of normal & mutant mtDNA will be received by daughter cell
|
|
|
IS THERE ANY EVIDENCE OF COOPERATION BTWN MITOCHONDRIAL & NUCLEAR GENOMES? |
Yes - Protein subunits (gene products) synthesized from Nuclear DNA pairing up with protein subunits from Mitochondrial DNA |
|
|
IS MAJORITY OF DNA TRANSCRIPTIONALLY ACTIVE OR INACTIVE? |
INACTIVE
Most of the DNA in the chromain is in form of highly compacted, highly methylated, DNase-resistant heterochromatin |
|
|
What base pair is usually methylated in DNA? |
Cytosine |
|
|
What are HATs and HDACs? |
HAT = Histone Acetyltransferases HDAC = Histone Deactylases
*Lysine is typically the A.A. involved
When Histone is Acetylated, it has no charge, and thus lower electrostatic affinity for DNA --> ENHANCED TRANSCRIPTION
When Histone is Deacetylated, it is + charged and has high affinity for DNA --> REPRESSED TRANSCRIPTION |
|
|
CpG island |
CpG island = Cytosine* - Phosphate Linkage - Guanine
Often found at the 5' regulatory regions of genes, involved in DNA methylation
*Cytosine receives the methyl group |
|
|
DNA methyl transferase |
Enzyme that methylates the Cytosine in DNA Methylation |
|
|
DNA Methylation - associated with Silenced transcription or Active Transcription? |
DNA METHYLATION --> SILENCED TRANSCRIPTION |
|
|
What factors are associated w/causing exogenous DNA Damage? |
1. UV Light exposure 2. Ionizing radiation
|
|
|
What factor is associated w/causing spontaneous DNA Damage? |
Cellular Metabolism |
|
|
What factor is associated w/causing DNA Damage as a result of replication errors? |
Chemical Exposure |
|
|
What problems w/Cellular Metabolism might cause SPONTANEOUS DNA DAMAGE? |
1. Deamination 2. Base Loss 3. ROS - Reactive Oxidative Species |
|
|
How does Deamination result in Spontaneous DNA Damage? |
*Involves spontaneous loss of an amine (NH3) -
-can result in formation of a non-standard base like Uracil, Hypoxanthine, Xanthine)
-or changes in Pyrimidines or Purines
|
|
|
How does Base Loss result in Spontaneous DNA Damage? |
*Spontaneously occurs at rate of 10,000 per cell per generation |
|
|
How do Reactive Oxygen Species (ROS) result in Spontaneous DNA Damage? |
H2O2 & Oxygen can contain/generate oxygens with unpaired electrons (Free Radicals) which can damage DNA |
|
|
What 2 vitamins may have protective effects against ROS? |
Anti-oxidants Vitamin C & E |
|
|
How does Ionizing Radiation result in (Exogenous) DNA Damage? |
*Ionizing Radiation = Gamma or X-rays
*Mediated via hydrolysis of water which breaks down into ROS |
|
|
How does UV light/radiation result in (Exogenous) DNA Damage? |
*DNA absorbs UV light (260nm)
*Can photoactivate nucleotides causing distortions in DNA helix
*Most reactive nucleotides are adjacent pyrimidines which become covalently linked to form PYRIMIDINE DIMERS
|
|
|
What are Pyrimidine Dimers? |
*Most commonly, Pyrimidine Dimers formed are Thymine dimers - Adjacent T's link together & create a cyclobutane - This formation distorts structure
*Bad b/c they prevent normal base pairing during subsequent replication |
|
|
How do Carcinogens cause (exogenous) DNA Damage? |
There are several types:
1. Aduct Formers (cigarettes) 2. Alkylating Agents 3. Cross Linking Agents |
|
|
What are Aduct Formers? |
Carcinogens that covalently attach to DNA nucleotides.
-Bulky -Interfere w/replication & transcription -Can lead to Base Substitutions
e.g. BENZO[A]PYRENE (Cigarette Smoke) |
|
|
What are Alkylating Agents? |
Carcinogens that are electrophilic chemicals
-They form covalent bonds w/electron rich atoms in DNA
-Usually involves adding methyl, ethyl or other large entity to one of the DNA bases, which disrupts stucture & pairing |
|
|
What are Cross Linking Agents? |
Carcinogens that can bond to 2 positions on DNA strand, forming crosslinks either inter or intra-strand
-The ones that facilitate inter-strand cross-links are most harmful b/c it inhibits replication & transcription completely |
|
|
What therapy/treatment option is associated w/Ionizing Radiation? |
Gamma irradiation |
|
|
What therapy/treatment option is associated w/Alkylating Agents? |
Cytoxan |
|
|
What therapy/treatment option is associated w/Crosslinking Agents? |
Cis platin |
|
|
What are Type II topoisomerase inhibitors? |
Type II topoisomerases cut both strands of the DNA helix simultaneously in order to manage DNA tangles and supercoils.
Type II topoisomerase inhibitors poison that enzyme & prevent it from working. They are often used as anticancer & antibacterial therapies *e.g. Etoposide inhibits re-ligation of DNA strands; induces double strand breaks
|
|
|
What Errors in DNA Replication commonly lead to DNA Damage? |
1. Polymerase Error 2. DNA Base Tautomers 3. Microsatellite Instability |
|
|
What is meant by "polymerase error?" |
Misincorporation of a nucleotide by polymerase (occurs in 1/1,000,000 bases)
That frequency is further reduced by "proofreading" and 3'-->5' exonuclease activity |
|
|
What are DNA Base Tautomers? |
Alternate forms of standard bases
*Rare, but the consequences are severe if happens during replication |
|
|
What is "Microsatellite Instability?" |
Occurs during Replication of repetitive DNA (known as microsatellites)
Can be thought of as "Slippage" of the primer strand relate to the DNA being copied -Produces mismatches or bulges in DNA -May lead to deletion/expansion of repeated sequences |
|
|
Possible Consequences of Damaged Genome on Cell Replication |
1. If DNA backbone IS intact -- replication can be error prone --> Translesion Synthesis
2. If DNA backbone is NOT intact -- replication can result in Double Strand Break, which is more difficult to repair -Mitosis in the setting of a double strand break can cause aberrant chromosome segregation (end up w/aneuploidy or translocations/deletions) |
|
|
What might happen at the cell cycle checkpoints in response to DNA Damage? |
Cell Cycle Arrest: Halting normal progression of cell in S or M phase to give it more time to repair damage
Cell Death: via Apoptosis, to remove cells that have acquired deleterious mutations |
|
|
DNA Repair Pathways & their Primary Targets |
1. BER = Base Excision Repair Targets: Deamination, Depurinations
2. NER = Nucleotide Excision Repair Targets: UV Photoproducts, Adducts, Cross-links
3. Translesion Synthesis (TS) Bypasses the Above, It is a tolerance process that allows the DNA replication to 'skip' past DNA lesions such as thymine dimers
4. Mismatch Repair (MMR) Targets: Replication errors
5. Homologous Recombination (HR) Targets: Dbl Strand Breaks, Adducts, Cross-links
6. End Joining Targets: Dbl Strand Breaks |
|
|
EXCISION REPAIR MECHANISMS |
*include BER, NER, MMR
*They excise damage on 1 strand, using undamaged complimentary strand as a template for repair
*very specific |
|
|
DOUBLE STRAND BREAK REPAIR |
*includes HR, End Joining
*They can't use a complementary strand for a template - that is why double strand breaks are considering the most deleterious form of DNA damage
*less specific, lower fidelity processes than Excision Repair |
|
|
How does Base Excision Repair (BER) work? |
Consists of recognition & repair of single-base alterations that occurred as a result of Deamination
-Employs specific DNA glycolysases (i.e. Uracil glycosylase for Uracil) and its specifity makes it the least flexible |
|
|
How does Nucleotide Excision Repair (NER) work? |
*Primary repair mechanism for UV photoproducts but can also repair damaged bases not recognized by a specific glycosylase
- Employs a large multiprotein complex (>10 proteins). Primary element of an active NER complex is XPA.
-NER recognizes damage and cuts 5 bases 3' of the damage....then cuts 23 bases 5' of the damage. The intervening ssDNA (~28 bases long) is then removed.
-Gap filled by DNA pol & ligase |
|
|
What is XPA? |
Xeroderma Pigmentosum group A gene product |
|
|
What diseases are associated w/defects in NER repair? |
1. Xeroderema Pigmentosum
2. Cockaynes Syndrome, Trichothiodystrophy |
|
|
What is Xeroderma Pigmentosum (XP)? |
Patients are sensitive to UV rays from sunlight & more susceptible to skin cancer
*Due to defects in NER pathway genes needed for transcription-coupled repair & global genome repair
*1:1,000,000
|
|
|
What is Cockaynes Syndrome/Trichothiodystrophy? |
Patients may be sensitive to UV, and NER compromised - but not always
*Developmental defects, hearing loss *Due to mutations in transcription-coupled NER or general defects in transcription |
|
|
Features of Transcription Coupled Repair |
1. NER damage recognition usually coupled to transcription 2. NER of damage in actively transcribed regions & in template regions is MORE EFFICIENT than non-transcribed "silent" regions 3. RNA polymerase encounters DNA lesion, stop and initiates formation of an NER complex. That NER complex repairs damage.
*Global Genome Repair - a transcription-independent mechanim that is less efficient, and slower |
|
|
TRANSLESION SYNTHESIS |
-not a true repair pathway, but still a way for replication to progress despite DNA damage
-synthesis bypasses damage in template strand
-occurs in a "non-templated" manner & is therefore error prone
-specialized DNA polymerases involved - pol eta, zeta, and mu |
|
|
What is Ataxia-telangiectasia (AT)? |
-variable autosomal recessive disease associated w/immune deficiencies, cerebellar ataxia, and telangiectasias of the conjunctiva (easy to spot in Ocular Exam, prominent blood vessels on the sclera)
-mutation in ATM gene (11q22.3)
-incidence of 1:40,000
-pts. are highly sensitive to irradiation & prone to lymphomas/leukemias
|
|
|
What is ATM? |
The gene mutated in Ataxia-telangiectasia (AT)
It is a large gene w/66 exons & 150kb that codes for a 3056 amino acid protein which regulates cellular responses to DNA damage |
|
|
DNA Mis-Matching ("Slippage") |
1. Backward slippage -- INSERTION
2. Forward slippage -- DELETION
Mis-matching more likely to occur in regions w/highly repetitive sequence |
|
|
DNA Mis-match Repair |
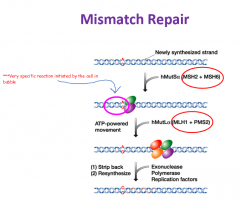
|
|
|
What is Hereditary Non-Polyposis Colon Cancer (HNPCC)? |
*aka Lynch Syndrome
*disease associated w/defective mismatch repair
*early (but variable) age of diagnosis ~45 yrs
*tumor site: proximal colon
*also associated w/extracolonic cancers, particularly endometrium & ovarian |
|
|
What are the Genetic characteristics of HNPCC? |
-Autosomal dominant inheritance -Genes belong to DNA mismatch repair (MMR) family -Genetic heterogeneity -Most commonly attributed to defects in MSH2 (30%) & MLH1 (30%) -MSH6 mutation (6%) as the cause is much rarer |
|
|
How can molecular pathology help us detect Colorectal cancers? |
Microsatellite Instability (MSI) Analysis: Would show shift peaks that are abnormal & indicative of high instability |
|
|
MSI Classification in Colorectal Cancers (CRC) |
-microsatellite stable (MSS)
-microsatellite instability low (MSI-L)
-microsatellite instability high (MSI-H) |

